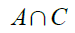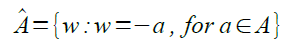In this activity, we were task in implementing past activity knowledge into object detection.segmentation. We were given two images shown in Figure 1 and 2.
Figure 1. Normal sized cells
Figure 2. Normal sized cells with five Abnormal Sized cells marked red
In figure 1, we are to assume each punched paper as normal cells. As can be seen, some of the cells overlap in pairs and in groups. For the first image, the goal would be to determine the best estimate of pixel area of one cell represented in terms of mean and standard deviation. Using these values, the abnormal sized cells in Figure 2 are to be isolated.
Thus the first step to be taken was to perform a threshold on the image. Analyzing the histogram of the image, thresholds were tested for values ranging from 170-210 and it was determined that 200 would be the best choice based on the result. As can be seen, the thresholded image has kept most of the cells circular in shape though a large part of the background was also detected in the right side of the picture, this will be removed later on after Morphological Operations.
Figure 3. Histogram of Figure 2.
Figure 4. After application of threshold
Given that the object of interest is circular, then the ideal Structuring Element should also be circular. Open function should therefor be implemented to filter out the background pixels via erosion while reconstructing the circular blobs through Dilation. Performing this, most of the imperfect cells, background image, as well as most of the cell outline are removed. In this activity, a circle structure element of size 12 was implemented.
Figure 5. (Left) Image after applying OpenImage. (Middle) Filtered out part of Figure 4. (Right) Detected cells in original image
FilterBySize() function was then used as a precautionary measure to remove any unwanted background pixel that wasn't filtered out; minimum pixel size was set to 100. Note that before this was applied, SearchBlobs() function was applied first. Thus each individual blob is marked by a separate value, summing the number of pixels corresponding to each blob value will give the pixel area of each blob. Doing this for the result in Figure 5, I was able to determine the pixel area of all 49 detected blobs in the image. Based from Figure 5, it can be seen that there are 10 blobs with overlapping cells, thus this cells should have a noticeable enlargement in pixel area compared to the other blobs corresponding to singular cells. This is shown in Figure 7, comparing the histogram with the image shown it can be assumed that blobs with pixel size greater than 600 do not belong to the singular cell group. Applying this via FilterBySize() function proves accurate. Thus calculating the mean and standard deviation of the pixel size shows that mean, µ = 498.35 pixels and standard deviation, σ = 24.28 pixels. Based from this the range of values corresponding to a normal sized cell is (µ
+/- 3*σ) pixels or 425 - 571 pixels. Based from this overlapping pairs of pixels can be assumed to be twice the pixel area of an individual normal sized cell, 600 - 1000 pixels. While overlapping cells in groups of three or more should be greater than 1000. Which can be seen to apply for the detected blobs as shown in Figure 9.
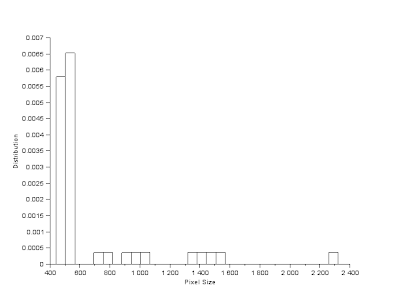
Figure 6. Histogram of Pixel Area of all detected blobs
Figure 7. Marked overlapping cells
Figure 8. (Left) Histogram of values corresponding to individual normal cells. (Right) Detected individual normal cells
Figure 9. After applying FilterBySize(Image, min, max). (Left) min = 600, max = 1000. (Right) min = 1000, max = infinity
Applying the same procedure for the image in Figure 2, filtering out the individual cells via FilterBySize() function by setting the minimum size as 571, I was able to get the abnormal sized cells along with overlapping cells. By filtering out overlapping cells in groups of three or more, only the abnormal size cells and the overlapping cell pair remains. Analyzing the histogram, it was observed that the pixel area of the abnormal sized cells corresponds with overlapping cell pairs. Thus the only other way of removing the overlapping pair from the image, aside from recreating a more complex morphological operation, was to implement a larger circular structure element such that normal sized cell would not be able to fit but abnormal sized cells would. Thus a circular structure element of size 13 was implemented.
Figure 10. After applying FilterBySize(Image, min, max). (Left) min = 571 (Right) min = 571, max = 1000
Figure 11. (Left) After applying new Structure element (Right) Inverting detected blobs and superimposing on original image
As can be seen the abnormal sized cells were isolated. In this activity, it was advised that the image in Figure 1 be divided into subimages and average the pixel area of the detected blobs to act as the average normal size of an individual cell. But this was determined via a different method in this activity. Nonetheless, I would like to give myself a grade of 10 for being able to complete the Activity and being able to understand what has been done. I am aware that I could done a better job at separating the blobs if a more thorough implementation of morphological operation was applied.
References:
M. Soriano, "Activity 11 - Application of Binary Operations 1". Applied Physics 186 2013. NIP, University of the Philippines -Diliman.